- Researchers developed a cost-effective way to collect DNA from species high in the rainforest canopy: they hung umbrellas to collect rainwater that washed through the trees.
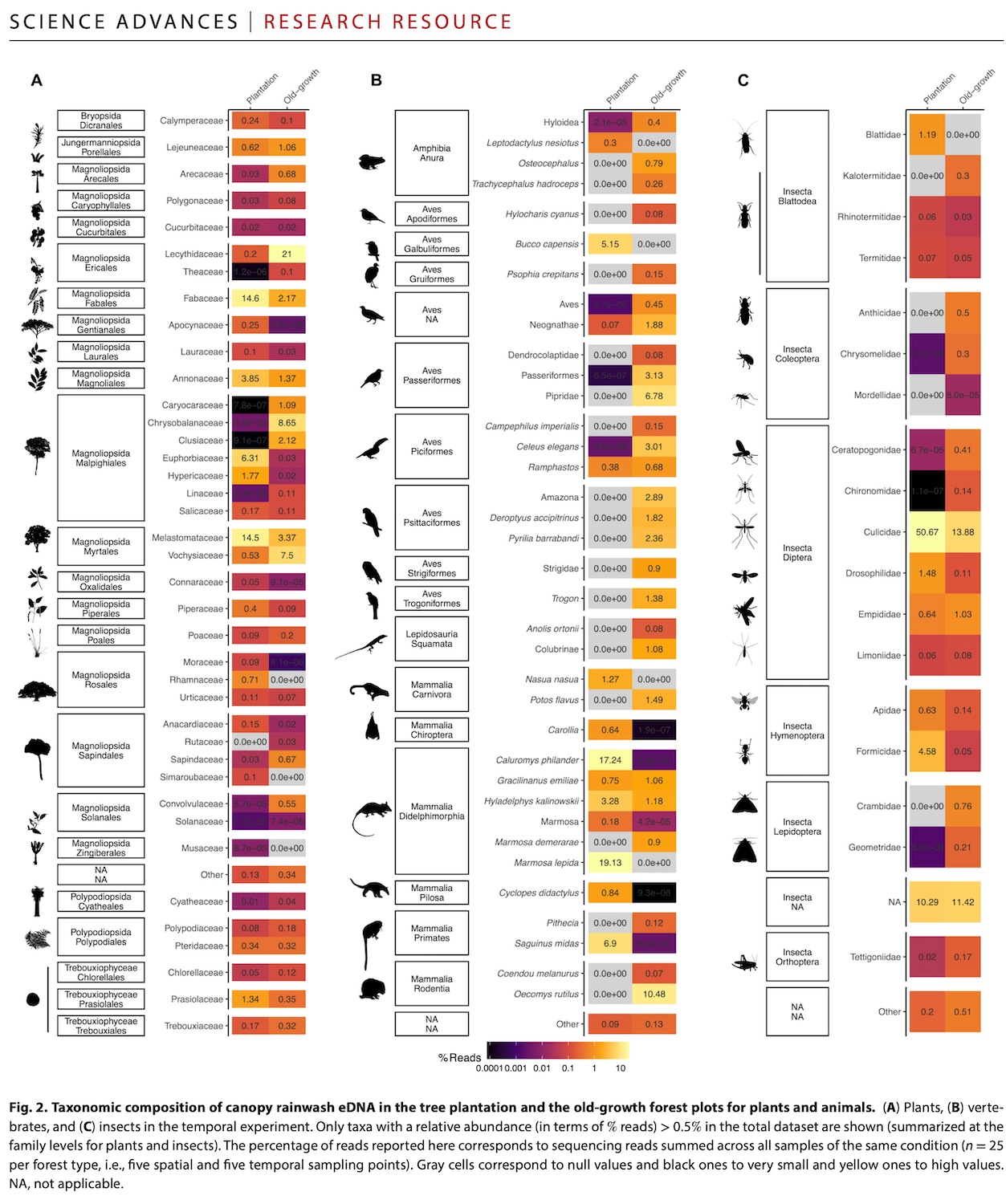
- The method revealed 562 species across French Guiana’s Amazonian forests, capturing genetic signatures from elusive nocturnal mammals, poorly documented reptiles, and countless undescribed insects that traditional survey techniques consistently overlook.
- Comparative analysis showed old-growth forests harbored 1.3 to 1.9 times greater species diversity than in nearby managed plantations.
- This simple technique provides local communities and conservationists with a practical way to monitor their forests, as each raindrop carries genetic evidence of the species present in the area over time.
In the rainforest, much of life dwells in the tree canopy. But getting up there to study it isn’t easy. By the time a human has clumsily lumbered up from the ground, most of the critters have scattered. So a team of researchers had an idea: Why not collect the rainwater that trickles down through the trees and analyze the water for traces of DNA?
Every living thing, from the tiniest moss to the largest monkey, sheds microscopic pieces of itself. Cells slough off. Saliva droplets land on leaves. Waste products get deposited on branches. When rain falls through the forest canopy, it washes all this biological debris downward, creating what scientists call rainwash, a carrier of environmental DNA, or eDNA.
To gather the rainwash, researchers repurposed ordinary umbrellas, drilling holes and attaching tubes to funnel water into collection devices, and hung them under trees in the Amazonian forests of French Guiana.

“We wanted to develop something low cost and easily usable for all the local actors for conservation,” Lucie Zinger, a researcher at the France-based Center for Biodiversity and Environmental Research (CBRE), told Mongabay. “So, the idea of the umbrella comes from something cheap and easy to use.”
The resulting study, published in Science Advances, found that the rainwater eDNA method detected a remarkable diversity of life in the canopy: 562 different species across plants, vertebrates and insects.
Testing two worlds
Researchers tested this rainwater method in the municipality of Sinnamary, at two contrasting sites: a 1-hectare (2.5-acre) plot of old-growth Amazonian rainforest at the Paracou Research Station, and a nearby rubber and rosewood plantation located 5 kilometers (3 miles) away.
The DNA signatures revealed creatures that have long evaded traditional research methods in tropical forests: secretive mammals that move through the canopy under cover of darkness, reptiles whose distributions remain poorly mapped, and the countless insects whose very existence often goes unrecorded by science.
Among the finds were Neotropical monkeys like tamarins and saki monkeys (from the genera Saguinus and Pithecia, respectively), two-toed sloths (genus Choloepus), anteaters, bats, and raccoon-like procyonids such as kinkajous (Potos flavus) and coatis (Nasua nasua). The plant diversity was equally remarkable, spanning 88 families from mosses and ferns to flowering trees typical of old-growth forests.
The differences between the two sites were clear. The old-growth forest harbored 1.3 to 1.9 times more species than the plantation.


In the forest, the tubes held some unexpected discoveries. “[We were] surprised to find fish DNA, likely from a bird’s diet, and also DNA from a house cat,” Amaia Iribar, CBRE researcher and co-author of the study, told Mongabay. The house cat’s DNA probably came from a cat wandering into the tree rom a nearby town.
Unlike traditional eDNA surveys that capture only a moment in time, like a scoop of water from a river, or a patch of soil, the passive rainwater collectors integrated biological signals over a period of eight to 40 days, depending on rainfall and DNA concentrations. This longer time frame provides a more comprehensive picture of an ecosystem’s inhabitants.
Unlike river water eDNA that can travel for kilometers, rainwater also provides a highly localized snapshot, capturing life from just tens of meters around each collection point. This precision allows researchers to pinpoint exactly where different species live and how human activities affect specific forest patches.
“We were really trying to have this passive collection approach, because we know that we can leave something at the field, and the [DNA] signal can accumulate somehow,” Iribar said. “And so that gives you a picture that is less temporary, a better description of local biodiversity.”

The challenge of the unknown
The precision of species identification varied dramatically across different groups of life. Plants achieved the finest resolution, with 56 of 88 families identified to species level, including mosses, ferns and the characteristic old-growth forest trees.
Vertebrates proved more challenging, with most identified only to genus or family level. However, the method still detected 21 vertebrate orders and could distinguish groups like Neotropical monkeys (tamarins and sakis), sloths and various bats.
Insects presented the greatest challenge, with most classified only to family or order level (flies, beetles, ants, moths and butterflies). This reflects both the incomplete state of genetic databases for tropical arthropods and a stark reality: an estimated 80% of insect species in French Guiana remain scientifically undescribed, meaning their genetic signatures simply don’t exist in reference databases.
“We have a very poor identification success [rate] because the reference database are really incomplete for tropical systems,” Zinger said.

Making the canopy more accessible
Despite the limitations in identification, the technique holds significant appeal for conservation practitioners in French Guiana.
“They are interested in these approaches because they want to see if their practices are actually successful,” Zinger said. “Practitioners in French Guiana were excited to detect reptiles for which there is a very poor descriptions and how they distribute and whether they are disturbed by roads or particular infrastructure.”
The rainwater method offers something precious in biodiversity research: accessibility. Traditional canopy studies require expensive equipment, specialized training, and physical risk. The umbrella collectors, by contrast, can be deployed by local communities, conservation organizations, or researchers with minimal resources.

“I think this paper is a very interesting new addition to the growing literature on eDNA applications for biodiversity monitoring,” Philip Francis Thomsen, an eDNA expert and professor at Aarhus University, Denmark, who wasn’t involved in the study, told Mongabay.”Tropical rainforest canopies are notoriously difficult to investigate noninvasively due to their inaccessibility, yet they contain a very high number of species – particularly insects, many of which are probably un-described.”
As climate change and deforestation accelerate across the Amazon and forests globally, tools like rainwater eDNA monitoring could prove crucial for tracking ecosystem health and guiding conservation efforts. The method transforms every raindrop into a tiny biological messenger, carrying news from the world above.
Banner image of researchers and study authors Amaia Iribar (left) and Lucie Zinger (right) preparing passive eDNA sampler in the forest of French Guiana. Image courtesy of Finn Piatscheck, UMR EcoFoG, INRAE.
Scientists find unexpected biodiversity in an African river, thanks to eDNA
Citation:
Zinger, L., Benoiston, A.-S., Cuenot, Y., Leroy, C., Louisanna, E., Moreau, L., … Iribar, A. (2025). Elusive tropical forest canopy diversity revealed through environmental DNA contained in rainwater. Science Advances, 11(33), eadx4909. doi:10.1126/sciadv.adx4909
Liz Kimbrough is a staff writer for Mongabay and holds a Ph.D. in Ecology and Evolutionary Biology from Tulane University, where she studied the microbiomes of trees. View more of her reporting here.
FEEDBACK: Use this form to send a message directly to the author of this post. If you want to post a public comment, you can do that at the bottom of the page.